QSAR Modeling and Molecular Docking Analysis of Some Active
4.6
(146) ·
$ 0.50 ·
In stock
Description

QSAR modeling, molecular docking, ADMET prediction and molecular dynamics simulations of some 6-arylquinazolin-4-amine derivatives as DYRK1A inhibitors - ScienceDirect

A comparison between 2D and 3D descriptors in QSAR modeling based on bio‐ active conformations - Bahia - 2023 - Molecular Informatics - Wiley Online Library

Virtual screening of PEBP1 inhibitors by combining 2D/3D-QSAR analysis, hologram QSAR, homology modeling, molecular docking analysis, and molecular dynamic simulations
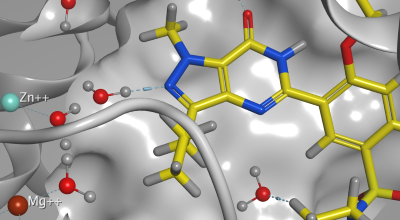
Molecular Operating Environment (MOE), MOEsaic
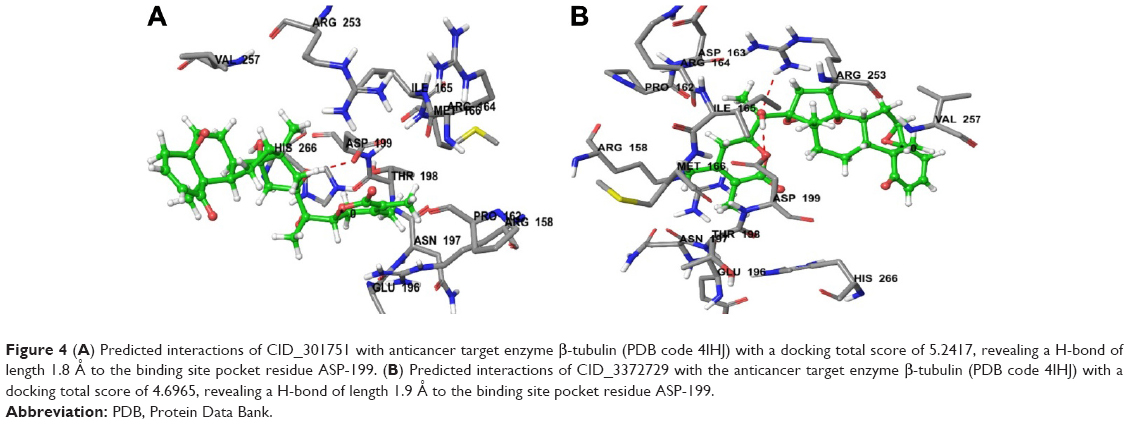
Molecular docking, QSAR and ADMET studies of withanolide analogs again

Novel QSAR Approach for a Regression Model of Clearance That Combines DeepSnap-Deep Learning and Conventional Machine Learning
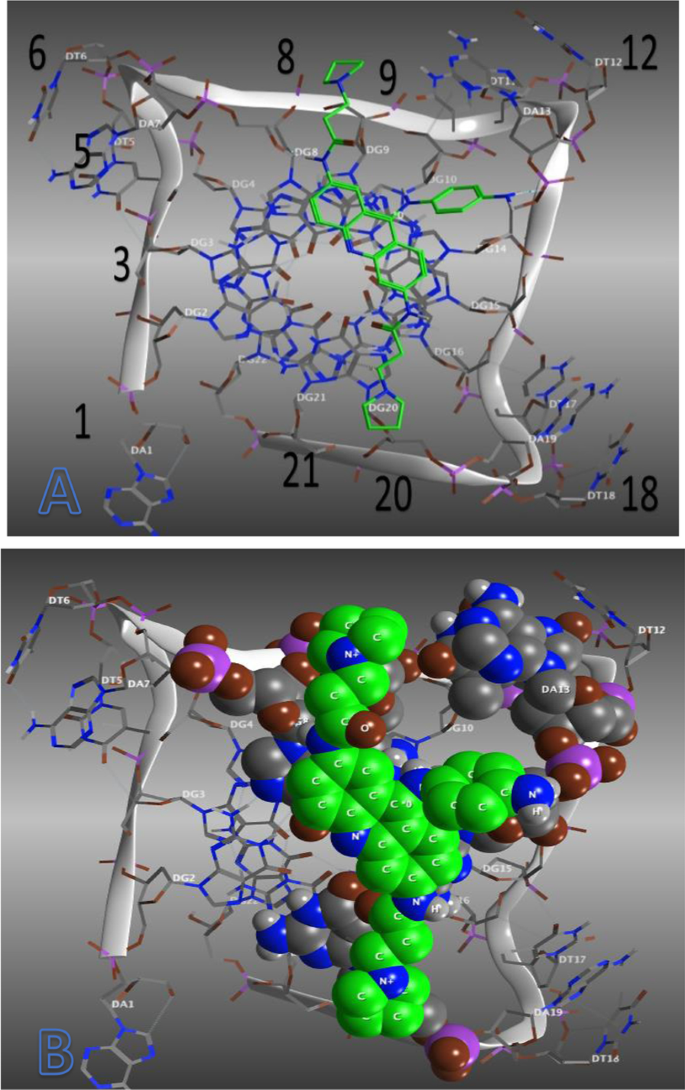
QSAR, pharmacophore modeling and molecular docking studies to identify structural alerts for some nitrogen heterocycles as dual inhibitor of telomerase reverse transcriptase and human telomeric G-quadruplex DNA
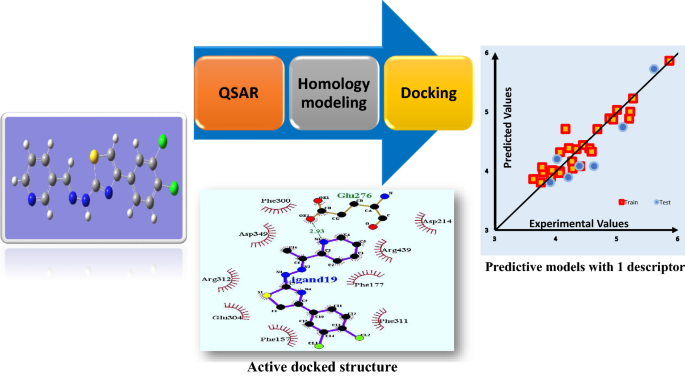
A simple and robust model to predict the inhibitory activity of α-glucosidase inhibitors through combined QSAR modeling and molecular docking techniques
Related products
You may also like
copyright © 2018-2024 lippe-wohnbau.de all rights reserved.






/cdn.vox-cdn.com/uploads/chorus_image/image/72350246/1252620395.0.jpg)
